Invariant Calculation
Principle
For any multi-phase system, i.e. any system that contains regions with different scattering length densities (SLD), the integral over all q (or 4π in scattering angle) of the appropriately dimensionally-weighted scattering cross-section (ie, ‘intensity’, I(q) in absolute units) is a constant directly proportional to the mean-square average fluctuation (SLD) and the phase composition. Usefully, this value is independent of the sizes, shapes, or interactions, or, more generally, the arrangement, of the phase domains (i.e. it is invariant) provided the system is incompressible (i.e, the relative volume fractions of the phases do not change). For the purposes of this discussion, a phase is any portion of the material which has a SLD that is distinctly different from the average SLD of the material. This constant is known as the Scattering Invariant, the Porod Invariant, or simply as the Invariant, Q∗.
Note
In this document we shall denote the invariant by the often encountered symbol Q∗. But the reader should be aware that other symbols can be encountered in the literature. Glatter & Kratky[1], and Stribeck[2], for example,both use Q, the same symbol we use to denote the scattering vector in SasView(!), whilst Melnichenko[3] uses Z. Other variations include QI.
As the invariant is a fundamental law of scattering, it can be used for sanity checks (for example, scattering patterns from the same sample that may look very different should have the same invariant if the hypothesis of what is going on in the sample is correct), to cross-calibrate different SAS instruments, and, as explained below, can yield an independent estimate of volume fractions or contrast terms.
Implementation
Calculation
Assuming isotropic scattering, acquired on a typical ‘pinhole geometry’ instrument, the invariant integral can be computed from the 1D reduced data as:
Warning
SasView, and to our knowlege most, if not all, other software implementations of this calculation, does not include the effects of instrumental resolution on the equation above. This means that for data with very significant resolution smearing (more likely to be encountered with SANS than with SAXS data) the calculated invariant will be distorted (it will be too high).
In the extreme case of “infinite” slit smearing, the above equation reduces to:
where Δqv is the slit height, and should be valid as long as Δqv is large enough to include most, if not all, the scattering. This limit is applicable, for example, to most data taken on Bonse-Hart type instruments. SasView does implement this limit so that, in contrast to above, the invariant calculated from such slit-smeared data could be more accurate than for normal pinhole SANS data which typically has sigificant Δq/q.
The slit smeared expression above has also been used to compute the invariant from unidirectional cuts through 2D scattering patterns such as, for example, those arising from oriented fibers (see the Crawshaw[4] and Shioya[5] references). However, in order to use the Invariant analysis window to do this, it would first be necessary to put the cuts in a data format that SasView recognises as slit-smeared by properly including the value of qv in the data file.
Note
Currently SasView will only account for slit smearing if the data being processed by the Invariant analysis are recognized as slit smeared. It does not allow for manually inputing a slit value. Currently, only the canSAS *.XML and NIST *.ABS formats facilitate slit smeared data. The easiest way to include Δqv in simple ASCII column data in a way recognizable by SasView is to mimic the *.ABS format. The data must follow the normal rules for general ASCII files but include 6 columns. The SasView general ASCII reader assumes the first four columns are q, I(q), dI, σ(q). If the data does not contain any dI information, these can be faked by making them ~1% (or less) of the I data. The fourth column must contain the qv value, in Å-1, but as a negative number. Each row of data should have the same value. The 5th column must be a duplicate of column 1. Column 6 can have any value but cannot be empty. Finally, the line immediately preceding the actual columnar data must begin with: “The 6 columns”. For an example, see the example data set 1umSlitSmearSphere.ABS in the \test\1d folder).
Data Extrapolation
The difficulty with using Q∗ arises from the fact that experimental data is never measured over the range 0≤q≤∞ and it is thus usually necessary to extrapolate the experimental data to both low and high q. Currently, SasView allows extrapolation to a fixed low and high q such that 10−5≤q≤10 Å-1.
Low-q region (<= qmin in data):
- The Guinier function I0.exp(−q2R2g/3) can be used, where I0 and Rg are obtained by fitting the data within the range qmin to qmin+j where j is the user-chosen number of points from which to extrapolate. The default is the first 10 points. Alternatively a power law, similar to the high q extrapolation, can be used but this is not recommended! Because the integrals above are weighted by q2 or q the low-q extrapolation generally only contributes a small proportion, say <3%, to the overall value of Q∗.
High-q region (>= qmax in data):
- The power law function A/qm is used where the power law constant m can be fixed to some value by the user or fit along with the constant A. m should typically be between -3 and -4 with -4 indicating sharp interfaces. The fitted constant(s) A (m) is/are obtained by fitting the data within the range qmax−j to qmax where, again, j is the user chosen number of points from which to extrapolate, the default again being the last 10 points. This extrapolation typically contributes 3 - 20% of the value of Q∗ so having data measured to as large a value of qmax as possible is generally much more important.
Note
While the high q exponent should generally be close to -4 for the assumptions underlying the extraction of other parameters to be correct, in the special case of slit smearing that power law should be -3 for the same sharp interfaces.
Invariant
SasView implements the invariant calculation for a two-phase (or pseudo two-phase) system, which represents the most commonly encountered situation. The invariant for this is
where Δρ=(ρ1−ρ2) is the SLD contrast and ϕ1 and ϕ2 are the volume fractions of the two phases (ϕ1+ϕ2=1). Thus from the invariant one can either calculate the volume fractions of the two phases given the contrast or, calculate the contrast given the volume fraction. However, the current implementation in SasView only allows for the former: extracting the volume fraction given a known contrast factor.
Warning
The Invariant analysis window always tries to return the volume fraction using a default SLD of 1e-6 Å-1. The user must provide the correct SLD for their system and click on Compute before examining/using the value of the invariant displayed.
Volume Fraction
In some cases, especially in non-particulate systems for which no good analytical model description exists (as then the scale factor of such a model would return the volume fraction information), if the contrast term can be reasonably estimated then the invariant can provide an estimate of the volume fraction. This is quite common, for example, in the Geosciences and Materials Science where the amount of porosity in a sample (the second phase) is of vital interest.
Rearranging the above expression for Q∗ yields
and thus, if ϕ1<ϕ2
where ϕ1 (the volume fraction of the minority phase) is reported as the the volume fraction in the Invariant analysis window.
Note
If A>0.25 then the program is obviously unable to compute ϕ1. In these circumstances the Invariant window will show the volume fraction as ERROR. Possible reasons for this are that the contrast has been incorrectly entered, or that the dataset is simply not suitable for invariant analysis.
Specific Surface Area
Warning
The value of Sv returned by this version 5.0.2 of SasView and ALL earlier versions of SasView is twice what it should be.
The total surface area per unit volume is an important quantity for a variety of applications, for example, to understand the absorption capacity, reactivity, or catalytic activity of a material. This value, known as the specific surface area Sv, is reflected in the scattering of the material. Indeed, any interfaces in the material separating regions of different scattering length densities contribute to the overall scattering.
For a two phase system, Sv can be computed from the scattering data as:
where Cp, the Porod Constant, is given by Porod’s Law:
which can be estimated from a Porod model fit to the an appropriately high-q portion of the data or from the intercept of a linear fit to the high-q portion of a Porod Plot: I(q)*q^4 vs q^4 (see the Porod model documentation in the Models Documentation for more details).
This calculation is unrelated to the Invariant other than to obtain the contrast term if it is not known (and the volume fraction is known), and depends only on two values - the contrast and Porod Constant - which must be provided.
Extension to Three or More Phases
In principle, as suggested in the Introduction, the invariant is a completely general concept and not limited to two phases. Extending the formalism to more phases, so that useful information can be extracted from the invariant is, however, more difficult.
We note here that in the more generalized formalism the contrast term is replaced by a quantity called the SLD fluctuation, \eta, so that:
where \eta represents the deviation in SLD from the weighted-average value, \langle (\rho^*) \rangle, at any given point in the system. The mean-square average of the SLD fluctuations, <\eta^2>, is:
In the case of a two-phase system:
Setting
then yields:
and thus:
Using invariant analysis
Load some data with the Data Explorer.
Select a dataset and use the Send To button on the Data Explorer to load the dataset into the Invariant panel. Or select Invariant from the Analysis category in the menu bar.
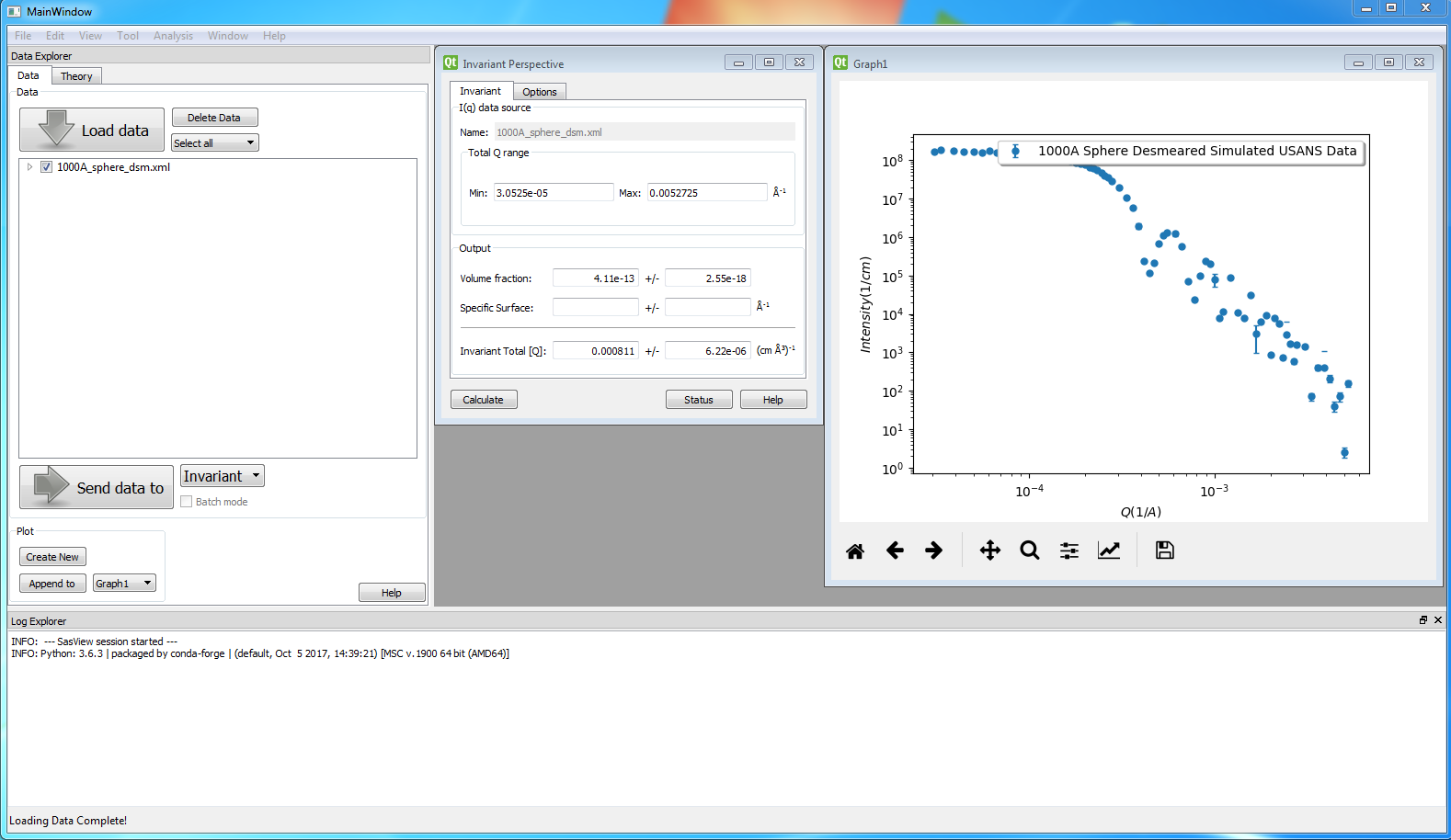
A first estimate of Q^* should be computed automatically. If not, click on the Compute button.
Use the boxes on the Options tab to subtract any background, specify the contrast (i.e. difference in SLDs: note this must be specified for the eventual value of Q^* to be on an absolute scale and to therefore have any meaning), or to rescale the data.
(Optional) If known, a value for C_p can also be specified.
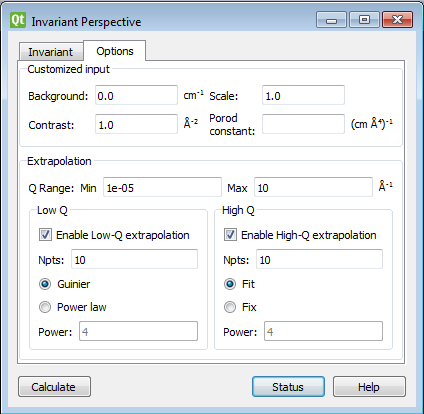
Adjust the extrapolation ranges and extrapolation types as necessary. In most cases the default values will suffice. Click the Compute button.
To adjust the lower and/or higher Q ranges, check the relevant Enable Extrapolate check boxes.
If power law extrapolations are chosen, the exponent can be either held fixed or fitted. The number of points, Npts, to be used for the basis of the extrapolation can also be specified.
The details of the calculation are available by clicking the Status button in the middle of the panel.
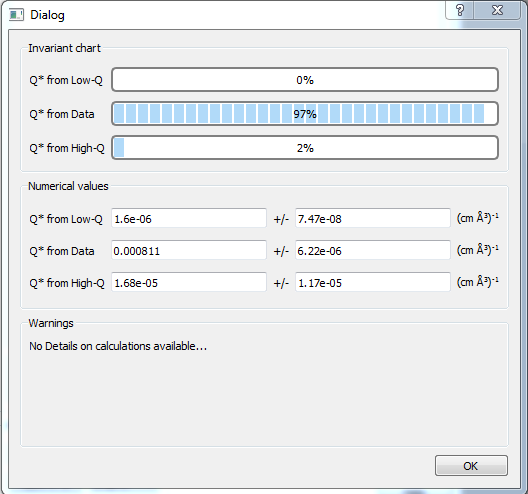
If more than 10% of the computed Q^* value comes from the areas under the extrapolated curves, particularly the high-Q extrapolation, proceed with caution.
References
| [1] | O. Glatter and O. Kratky Chapter 2 and Chapter 14 in Small Angle X-Ray Scattering; Academic Press, New York, 1982. Available at: http://web.archive.org/web/20110824105537/http://physchem.kfunigraz.ac.at/sm/Service/Glatter_Kratky_SAXS_1982.zip. |
| [2] | N. Stribeck Chapter 8 in X-Ray Scattering of Soft Matter Springer, 2007. |
| [3] | (1, 2) Y.B. Melnichenko Chapter 6 in Small-Angle Scattering from Confined and Interfacial Fluids; Springer, 2016. |
| [4] | (1, 2) J. Crawshaw, M.E. Vickers, N.P. Briggs, R.K. Heenan, R.E. Cameron Polymer, 41 1873-1881 (2000). |
| [5] |
|
Note
This help document was last changed (completely re-written) by Paul Butler and Steve King, 16Apr2020